GROWTH PROPERTIES AND VIRUS STRUCTURE
In 1986, Salahuddin and coworkers1 isolated HHV-6 from peripheral mononuclear cells of patients with various lymphoproliferative diseases and from an individual with human immunodeficiency virus type-1 (HIV-1) infection. More than 10 laboratories worldwide have reported the isolation of closely related viruses.2
Because of its similarity to HCMV, HHV-6 is classified as a betaherpesvirus.2 Immunofluorescent staining of HHV-6-infected cultures using specific monoclonal antibodies to other known HHVs—HSV-1, HSV-2, VZV, EBV, and HCMV clearly indicate that HHV-6 is a previously undetected virus. In situ hybridization using cloned viral deoxyribonucleic acid (DNA) probes has further confirmed these findings.3 The HHV-6 isolates from several laboratories exhibit differences in in vitro cell tropism, effects on expression of T-cell markers, the major cell type that supports their growth, the restriction endonuclease patterns of their DNA, differential reactivity with various monoclonal antibodies, seroepidemiology, and disease associations.4,5 Based on these observations, HHV-6 is divided into two groups: A and B. The two variants of HHV-6 are genetically closely related.2–6 A clear etiologic role of HHV-6A has not been identified, although HHV-6B has been shown to be the major cause of exanthem subitum or roseola.7
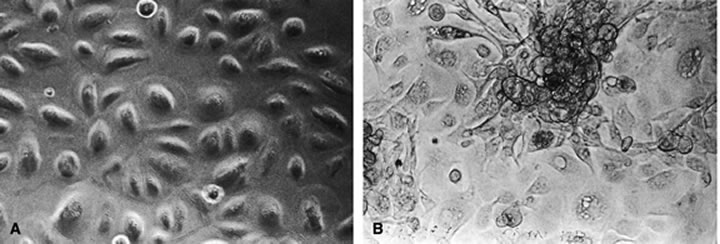
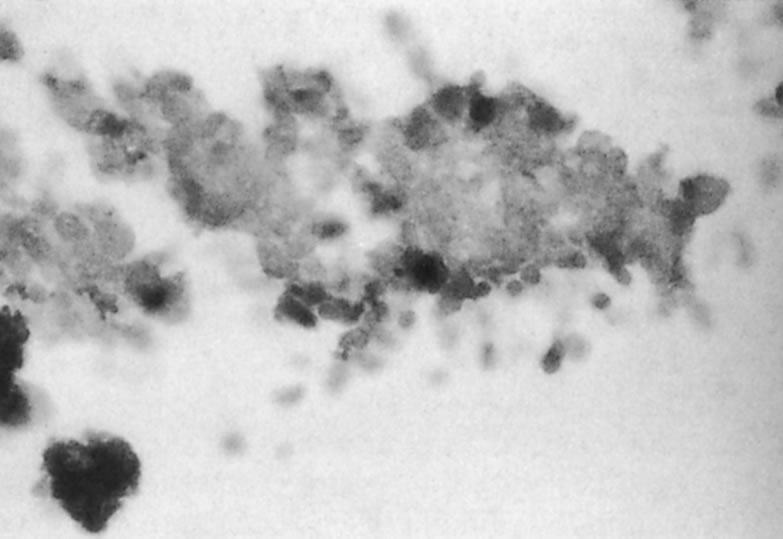
The predominant cell type permissive for replication of both HHV-6A and HHV-6B is CD4+ T cells. Although individual isolates of both variants A and B differ in their host range and ease of adaptability to culture, isolates of both kinds have been adapted by serial passages in continuous cell lines to yield higher titers. An often noted variant differentiating characteristic is the relative ease of adapting HHV-6A isolates for growth in T-cell lines, CCRF HSB2 cells and HHV-6B isolates in Molt 3 cells.6 A range of other cell types (including some EBV transformed B-cell lines, transformed cervical epithelial cells, megakaryocytes, glial cell lines, neurons, astrocytes, primary monocytes/macrophages, natural killer cells, bone marrow, tonsil, thymus, and spleen mononuclear leukocytes) have been infected with HHV-6. HHV-6B isolates have a more restricted cell tropism than the HHV-6A variant.2 HHV-6 has been recovered by cocultivation of corneas with peripheral blood monocytes, and the virus is capable of infecting primary and transformed corneal epithelial cells. HHV-6 can infect corneal cells, which then express viral antigens and DNA sequences.8 HHV-6 induces cytopathic effects in infected cells, including corneal cells (Fig. 1A and B). These changes are characterized by the presence of large balloon-shaped refractile cells, with occasional syncytia formation in the infected cultures. The cytopathic effects are visible 4 to 5 days after infection, and virus is usually cell associated.2,6,8 HHV-6 has also been isolated from and detected in retinas of acquired immunodeficiency syndrome (AIDS) patients9–11 (Fig. 2).
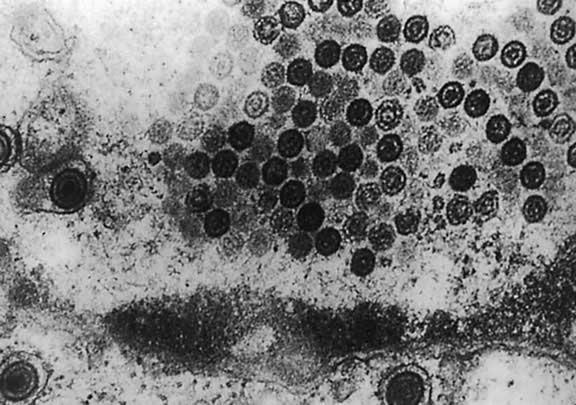
Electron microscopy shows that HHV-6 has features typical of members of the herpesvirus family. Its DNA is present in an icosahedral capsid made up of 162 capsomeres. The enveloped virions are 160 to 200 nm in diameter; naked capsid is 90 to 110 nm, and enclosed cores are 60 nm in diameter. Empty and DNA-filled nucleocapsids are observed in the nucleus. In the cytoplasm, capsids are coated with a fibrous material called tegument, which is 20 to 40 nm in diameter (Fig. 3). The tegument is the most striking morphologic feature of HHV-6; this component of the virion is more clearly demarcated in HHV-6 than in HCMV virions. Mature enveloped virions with prominent tegument accumulate in cytoplasmic vacuoles and on the surface of infected cells. Large inclusion bodies are also present in the cytoplasm of some infected cells.12,13
|
The double-stranded linear DNA genome of the virus has been estimated to be 163 to 170 kilobase pairs (kbp), with an overall G + C content of about 43%. The HHV-6 DNA consists of a 143-kbp unique long segment flanked on both sides by single copies of direct repeated (DR) sequences (i.e., in the same orientation). The size of the DR sequences is 13 kbp in HHV-6A strain and 10 kbp in HHV-6B strains.14 Less than 10% of the genome has been shown to be circularized, directly linking the left DR with the right DR.
Restriction fragment polymorphism has been noted when comparing different HHV-6 isolates, indicating variation in DNA structure among isolates.15 Restriction-enzyme analysis of purified HHV-6 DNA shows a pattern markedly different from other HHVs. Under stringent conditions, however, HHV-6 DNA cross-hybridizes to certain regions of HCMV DNA.16
In the HHV-6 genome, seven blocks in the central region are core genes, which are common to all herpesviruses. Another block spanning open reading frames (ORFs) U2 to U14 contains specific genes for the betaherpesvirus subfamily. The ORFs U15 to U25 are genes specific to roseolavirus (HHV-6 and HHV-7). However, three genes U22, U83, and U95 are specific to HHV-6 and are not found in HHV-7. HHV-6 also closely resembles HHV-7 in genome homology and organization. Amino acid similarity to HHV-7 is 46.6% to 84.9% and to HCMV 41.0% to 75.8%.15–19
REPLICATION, LATENCY, AND REACTIVATION
HHV-6 appears to enter cells by endocytosis. The nature of the receptor is unknown; however, monoclonal antibody to virion glycoprotein complex seems to inhibit the penetration of HHV-6 into the cells.19 The virus has been observed to enter uncoated pits, and the viral membrane fuses with endocytic vesicles, resulting in the release of nucleocapsids into the cytoplasm. The nucleocapsid enters the nucleus, where gene expression begins and viral DNA replication takes place. The nucleocapsid is assembled, acquires the tegument in unique nuclear structures called tegusomes, and is then released into the cytoplasm by fusion of the tegusome with the nuclear membrane. The resulting particles are enveloped by budding into cytoplasmic vacuoles, which then fuse with the cell membranes to release mature virions.13,20
The virus readily infects and replicates in some T-cell lines and fibroblasts in vitro. In other cell lines, however, virus replication is suppressed or the virus may become latent. Latent viral genome has been detected in T- and B-cell lines and fibroblasts that survive viral infection. The only data available to support latency is the failure to recover HHV-6 from peripheral blood lymphocytes of all healthy adults, indicating that the virus may be latent in these cells. During primary infection, HHV-6 DNA is detectable in nonadherent peripheral blood lymphocytes, whereas during convalescence it is only detectable in adherent cells by polymerase chain reaction (PCR). Several investigators have been able to reactivate HHV-6 from latently infected macrophages, from T- and B-cell lines and from fibroblasts using 12-0-tetradecanoylphorbol 13-acetate and hydrocortisone.2,21
VIRAL DNA REPLICATION AND GENE EXPRESSION
The temporal relation of DNA replication and viral gene expression is unknown. Little is known about the regulatory properties of HHV-6 promoters or the role of specific viral genes in HHV-6 gene regulation. HHV-6 codes for a variety of enzymes and regulatory proteins involved in nucleotide metabolism. Some of these have been identified as homologues of genes present in HSV-1, VZV, and HCMV, and others have been biochemically characterized.
These genes encode the major DNA-binding protein, DNA polymerase, polymerase-associated protein, a helicase-primase complex, DNA origin binding protein, and the small subunit of ribonucleotide reductase. Similar to HCMV, however, a virus-specific thymidine kinase activity has not been demonstrated in cells infected with HHV-6. The absence of a gene for thymidine kinase has also been confirmed by sequence homology analysis.15
Because of the low abundance of viral transcripts, many genes expected to be expressed during productive replication have not been identified by Northern blot analysis, in which the only transcript detectable was a 3.5-kb transcript that was identified as a product of the immediate early regulatory gene or genes. The C-terminal portion of this immediate early protein has the ability to transactivate heterologous promoters.22
With an estimated 170-kbp DNA, HHV-6 is capable of coding for more than 70 proteins. Using purified virions, more than 25 HHV-6-specific proteins, ranging from 30 to 200 kilodaltons (kDa), have been identified by immunoprecipitation. Among these, nine are glycoproteins, eight are associated with infected cell membranes, and six are associated with viral envelope. In addition to the previously mentioned proteins necessary for DNA synthesis, HHV-6 encodes genes for a large structural phosphoprotein, a large tegument protein, a major capsid protein, and an HHV-6-induced nuclear antigen.23 In addition, HHV-6 but not HHV-7 carries a homologue of adeno-associated type 2 parvovirus rep gene that is transcribed in latently infected cells.24,25 Recently, the viral protein from the U12 gene has been recognized as a beta-chemokine receptor.25 HHV-6 may play a role in tumor progression through these molecules.
EPIDEMIOLOGY AND PATHOGENESIS
Worldwide, more than 95% of individuals older than the age of 2 years are seropositive for HHV-6 antibody. The level of maternal antibody decreases during the first 5 months of life; antibody levels sharply increase between 6 months and 5 years of age, indicating initial infection. The seroprevalence decreases with age; however, it rises again at age 62 years. The mode of transmission is believed to be saliva. HHV-6 DNA has been detected in saliva of 90% of the United States population.2,27
HHV-6-associated exanthem subitum (a childhood disease) is characterized by high fever and a rash for 2 to 5 days. The incubation period is usually 5 to 15 days, and the disease normally resolves without complications. In rare cases, complications such as recurrent convulsions, meningitis, hepatosplenomegaly, mononucleosis, and fatal fulminant hepatitis have been reported.7,27–29 In the adult population, primary HHV-6 infection is rare. If infection is symptomatic, the symptoms can include lymphadenopathy, mononucleosis, and hepatitis.27
Virus-neutralizing IgM has been reported in patients 4 to 8 days after the onset of exanthem subitum, which usually peaks after 2 to 3 weeks and disappears in 7 to 8 weeks. In most cases, IgG is detectable after the eighth week. Cellular immunity may have a role in controlling HHV-6 infection in organ transplantation patients. The simple tests employed for the detection of HHV-6 antibodies (specifically, IgG levels) are indirect immunofluorescence and anticomplement immunofluorescence. The most widely used and most sensitive test is the enzyme-linked immunosorbent assay. The most sensitive and specific markers for HHV-6 infection are 135-kDa and 100-kDa antigens present in the serum.2
One intriguing characteristic of HHV-6 is its interaction with other viruses. It has been shown that HHV-6 grows with EBV or HCMV simultaneously within the same cells and may interact with each of these viruses. HHV-7 has been demonstrated to activate latent HHV-6. An enhanced expression of human papilloma virus (HPV) oncoproteins, E6 and E7, and transactivation of an HPV-18 promoter in cervical epithelial cells in the presence of HHV-6 infection has been documented, indicating that the virus may be associated with induction of cervical carcinoma caused by HPV.27 There are strong indications that HHV-6 may not transactivate human T-cell leukemia virus type I long terminal repeat.30 It has been shown that HHV-6 accelerates the destruction of CD4+ T lymphocytes in cultures coinfected with HHV-6 and HIV-1.8,31
HHV-6 infection induces nuclear factors that specifically bind to the enhancer region of HIV-1 LTR.31 When cloned into T- cells, HHV-6 DNA fragments containing an immediate early gene encode gene products that are able to activate the HIV-1 LTR.32 Because of its positive interaction with HIV-1, a catalytic role for HHV-6 during progression of AIDS has been proposed. The activation of HIV-1 by HHV-6 in retinal cells in vitro and the activation of HSV-1 in rabbit corneas in vivo have been demonstrated.8,33 These findings suggest that HHV-6 may play an important role in the pathogenesis of retinitis and keratitis.
The oncogenic potential of HHV-6 has been tested in mouse fibroblast cell lines and human epidermal keratinocytes in vitro using three viral DNA fragments.19 One codes DR7 and the other two encompasses the regions spanning U2 to U20 and U31 to U37. DR7 and the two other loci also contain genes with transactivating activity on HIV-1 LTR.34,35
HHV-6 is one of the most neurotropic viruses known in children and adults. A possible correlation between active HHV-6 infection and multiple sclerosis (MS) has also been the focus of much attention in the past few years. MS is a severe central nervous system (CNS) disease of young adults characterized by the progressive demyelination of nerves that lead to paralysis and eventually to death. This is an autoimmune disease of myelin. HHV-6B DNA sequences were found with higher frequency in MS brain specimens using representational difference PCR.36 Oligodendrocyte nuclei were found positive for HHV-6 antigens in MS samples (80%), whereas the controls were negative (0%).The titer of anti-HHV-6 IgM was also higher in samples from MS patients than control (73% vs. 8%).These findings are interpreted as a hallmark of the association between active or reactivable HHV-6 infection and the disease.37–39 The serologic analysis are difficult to interpret, because this disease is caused by an immunologic dysregulation and, therefore, the rise in antibody titer may be the sign of the disease rather than the cause. However, these results were not confirmed by other investigators.40 Therefore, the association between an active infection and MS is still a controversial issue.
Foscarnet, a potent inhibitor of DNA polymerases of other herpesviruses, has been shown to successfully inhibit HCMV and HHV-6 infections in AIDS patients with HCMV retinitis.41 Ganciclovir and foscarnet have been used to successfully treat HHV-6 infections in several organs transplant patients.42 Sophocarpines have been shown to be effective against HHV-6 replication (Qavi and coworkers, manuscript accepted by Phytotherapy for March 2002 issue [in press]).