BACTERIA
Most bacteria are not visible in unstained tissue. The standard initial staining method for identifying bacteria is Gram's stain. To perform a Gram stain, the sample is spread on a slide, dried, and briefly fixed in methanol. It is then stained with a cresyl violet solution (i.e., Hucker's solution) for 1 minute, rinsed in water, and then stained in Gram's iodide for 1 minute. Following decolorization in 95% ethyl alcohol and a brief water rinse, the sample may be counterstained with safranin O. The entire procedure can be completed in less than 5 minutes. Gram-positive organisms resist the decolorization step and remain stained dark purple, whereas gram-negative bacteria have their cell wall permeability increased by the alcohol wash and thus decolorize; they are visualized by the pink counterstain. Table 1 lists common gram-positive and -negative organisms causing ophthalmic disease.
Table 1. Common Gram-Positive and Gram-Negative Bacteria
Causing Ocular Disease
| Genus | Representative Species | Gram's Stain | Common Clinical Context |
| Staphylococcus | aureus, epidermidis | Positive cocci (clusters) | Cellulitis, keratitis, endophthalmitis |
| Streptococcus | pneumoniae, viridans | Positive cocci (chains) | Conjunctivitis, keratitis, blebitis, endophthalmitis |
| Bacillus | cereus | Positive rod | Traumatic endophthalmitis, keratitis |
| Clostridium | perfringens | Positive rod | Keratitis |
| Corynebacterium | ulcerans | Positive rod (club shaped) | Conjunctivitis, keratitis |
| Listeria | monocytogenes | Positive rod | Keratitis |
| Propionibacterium | acnes | Positive rod | Late onset endophthalmitis |
| Actinomyces | israelii | Positive rod (branched) | Keratitis, caniliculitis |
| Nocardia | asteroides | Positive rod | Keratitis, choroiditis |
| Neisseria | gonorrhoeae | Negative cocci | Conjunctivitis, keratitis |
| Branhamelia | catarrhalis | Negative cocci | Keratitis |
| Escherichia | coli | Negative rod | Keratitis |
| Haemophilus | influenzae | Negative rod | Blebitis, keratitis |
| Klebsiella | pneumoniae | Negative rod | Keratitis |
| Moraxella | catarrhalis | Negative rod | Keratitis |
| Proteus | mirabilis | Negative rod | Keratitis |
| Pseudomonas | aeruginosa | Negative rod | Keratitis |
| Serratia | marcescens | Negative rod | Keratitis |
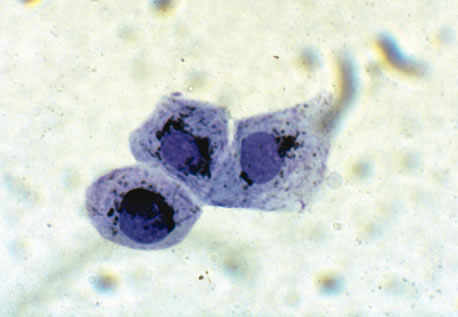
Certain bacteria are not readily visualized by Gram's stain, and may require other stains or types of microscopy for visualization. Mycobacteria are most easily visible using acid-fast stains such as the Ziehl-Neelsen stain. Intracellular Chlamydia are not directly visible with Gram's stain, and are generally visualized following Giemsa staining of intracellular inclusion bodies (Fig. 1). It is important to note that this Giemsa stain is the traditional 60-minute staining protocol; the brief Wright-Giemsa stain used for blood stains will not identify inclusion bodies. The Papanicolaou stain can also be used for the identification of Clamydia inclusions. Giemsa stain is also useful for the demonstration of Actinomyces and Nocardia species (which can also be seen on Gram's staining). Darkfield microscopy can be used to demonstrate the Treponema pallidum spirochete of syphilis, although serologic testing is more commonly performed to confirm infection. Serologic testing is more commonly performed to confirm the traditional 60-minute Giemsa staining protocol; the brief Wright-Giemsa stain used for blood stains will not identify inclusion bodies. The Papanicolaou stain can also be used for the identification of Chlamydia inclusions.
|
Certain direct fluorescent stains may be used in lieu of Gram's strain for detection of bacteria. In particular, acridine orange has a much higher sensitivity for detection of bacteria than Gram's stain, and has the additional advantage of staining fungi more reliably than Gram's stain. The drawback of these direct fluorescent stains is the necessity of a fluorescent microscope for detection of the fluorescent signal. Positive specimens should be secondarily strained with Gram's stain to identify the detected pathogen.
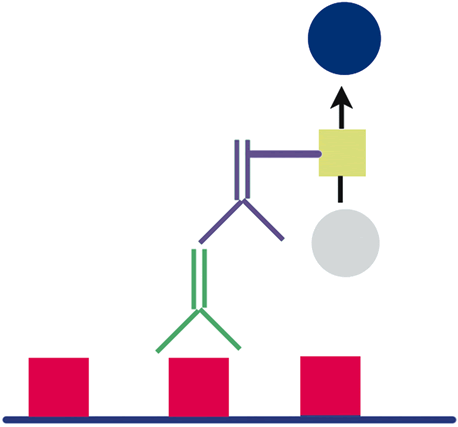
Commercial monoclonal or polyclonal antisera can be used to identify specific organisms by indirect immunofluorescence. In ophthalmology, this is commonly used for Chlamydia species.1 Because a specific antiserum must be used for each suspected organism, the requesting physician must communicate with the clinical microbiology or pathology laboratory in order to request these tests.
FUNGI
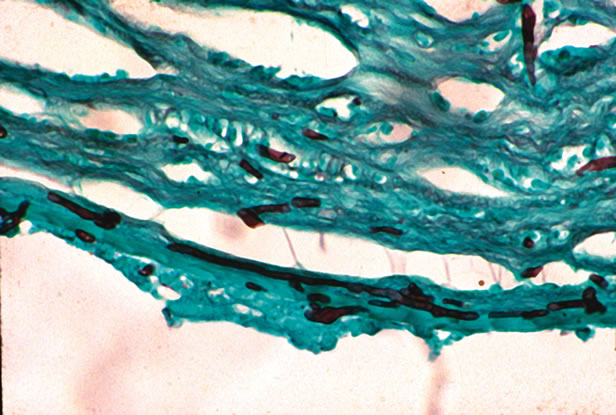
The most rapid technique for visualizing yeast or filamentous fungi is the potassium hydroxide (KOH) preparation. The scraping or swab is treated with 10% KOH in dimethyl sulfoxide. The strong base dissolves keratinized tissue (such as cornea), but the cell walls of fungi are resistant to dissolution. Both budding yeast and filamentous fungi can be visualized. Other staining methods also have high sensitivity for detection of yeast and fungi, including Giemsa stain, Gomori's methenamine silver (GMS), India ink staining, or periodic acid–Schiff (PAS) stain (Fig. 2). Fungi can also be identified on some Gram-stained samples.
|
VIRUSES
Viruses are typically submicrometer in size and cannot be directly visualized by light microscopy. Characteristic cellular changes can sometimes be identified as, for example, the cytoplasmic inclusion bodies of herpes infection on hematoxylin-eosin–fixed sections or on the Tzanck smear (in which the fluid from a suspected herpetic vesicle is stained with Giemsa, Papanicolaou, and Wright's stain; multinucleated giant cells indicate herpetic infection). Viruses can be directly observed by electron microscopy, but this is rarely used clinically because of the difficulty in specimen preparation and the low yield of positive results. Specific antisera for many viruses are available, and can be used to identify virally infected cells; however, this technique is typically used in research settings because it generally requires fixed biopsy tissue with preserved cytoarchitecture.
PARASITES
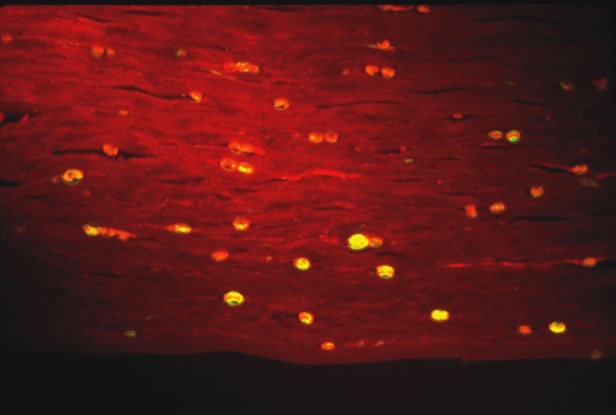
Some parasites can be seen free living under the microscope. Onchocerca volvulus, the causative agent of river blindness, can be directly observed swimming out of skin or corneal biopsies incubated in saline or culture medium. Modern imaging techniques are increasing the range of organisms that can be directly detected in situ. In particular, the confocal microscope has been successfully used to visualize Acanthamoeba in infected corneas.2 Certain parasites causing ophthalmic disease can be visualized with special stains. Acanthamoeba species can be visualized on corneal scrapings with PAS or GMS stains, or with the fluorescent dyes calcifluor white or acridine orange (Fig. 3). Both techniques require a fluorescent microscope to visualize the dye. Calcifluor white (a whitening agent used in laundry detergents) stains Acanthamoeba cell walls and fluoresces green under ultraviolet illumination. Toxoplasma gondii can be visualized with PAS or GMS stains from biopsy material. Specific antisera can be used in stains of biopsy specimens.
|