GENOME STRUCTURE AND GENE EXPRESSION
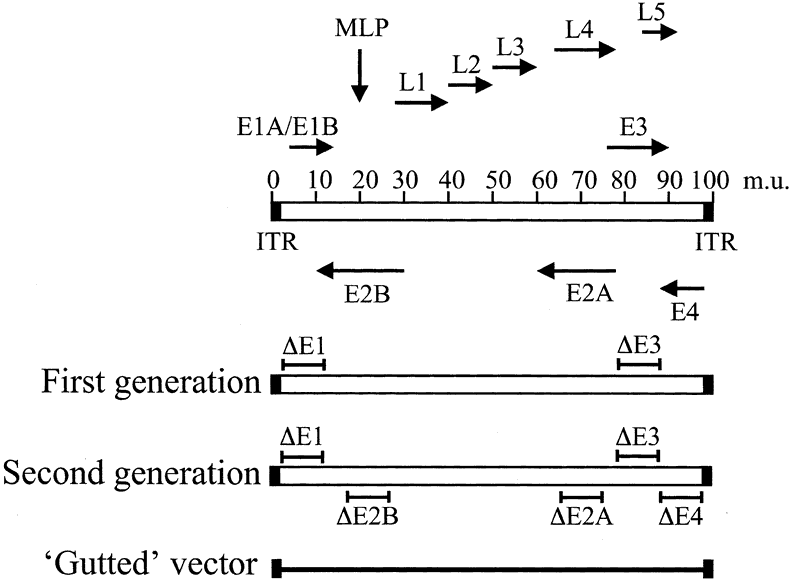
The 49 serotypes of human adenovirus have been subclassified into six subgroups (A through F).15 The human adenovirus serotypes 2 and 5 (Ad2 and Ad5, respectively), both from subgroup C, have been most extensively characterized and have been used recently for construction of viral vectors for gene therapy. (See the following section on adenovirus-mediated gene transfer.) Ad2 and Ad5 genomes have been completely sequenced and are 95% identical at the nucleotide level, with a similar organization of transcriptional units (reviewed in ref 22). The Ad5 genome is a linear double-stranded DNA of approximately 36 kilobases (kb), with at least 30 different messenger ribonucleic acids (mRNAs). The mRNAs may be grouped into early and late transcripts, according to whether they are expressed before or after viral DNA replication (Fig. 1). Early transcripts (E1A, E1B, E3, and E4) encode proteins that modulate the host cell cycle, mediate viral DNA replication, and control the host immune response to viral invasion.23 Late transcripts (termed L1 through L5) encode viral structural proteins such as hexon, penton base, and fiber. In the late phase of the viral cycle, the major late promoter (MLP) is activated and a large precursor mRNA is synthesized. The L1 to L5 transcripts are derived from this common transcript through alternative splicing. Other small transcripts that have important functions in the viral life cycle encode pIX (a structural protein), IVa2 (a protein that activates the MLP), and VA RNA I and II (RNAs that block activation of the host interferon response).
STRUCTURE OF THE VIRUS
Adenoviruses are nonenveloped viruses. The viral structural proteins are DNA-associated or involved in formation of the complex capsid encapsulating the viral DNA. The DNA-associated proteins include the terminal protein (TP) that binds to the 5' termini of the viral genome containing the inverted terminal repeats (ITRs).24 Other core proteins include polypeptides V and VII and Protein X. Like cellular histones, these proteins are rich in the basic amino acid arginine, but unlike histones they contain appreciable amounts of tryptophan.25,26 The precise function of each of the core proteins has yet to be elucidated, but protein V may play a role in interfering with host cellular function by redistributing nucleolar antigens.27
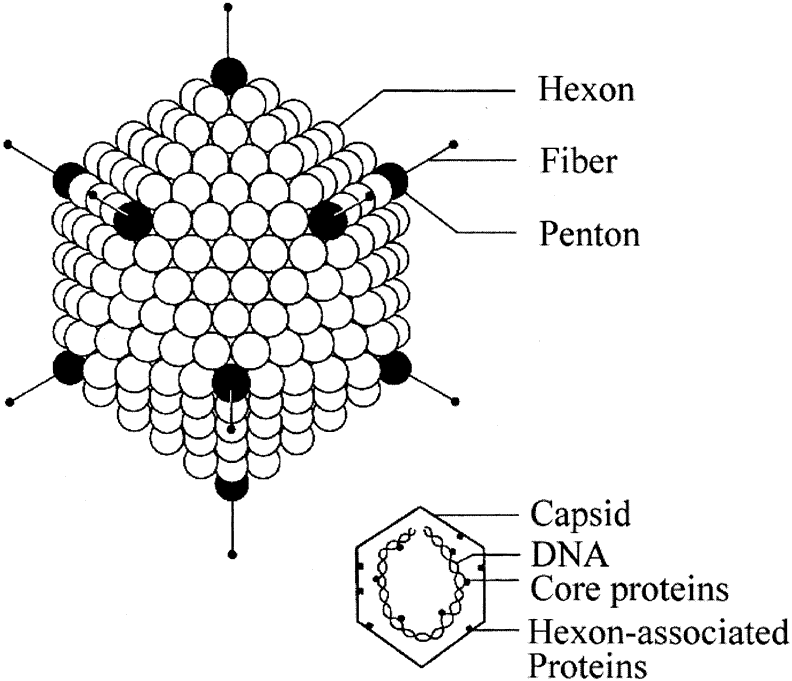
The icosahedral capsid structure of adenovirus is made up primarily of three proteins: hexon, penton base, and fiber as shown in Figure 2. The capsid is formed from 252 unit structures, known as capsomeres, arranged in an icosahedral structure with 20 triangular sides. Hexagonal capsomeres or hexons comprise 240 of the structural subunits.28 Each hexon is composed of three different polypeptides VI, VII, and IX. There are also 12 pentagonal capsomeres or pentons. Each penton is composed of an 85 kilodalton (kDa) protein, polypeptide III. Polypeptide III is the largest protein of the entire viral capsid. Projecting from each penton is a rodlike structure called a fiber made up of three molecules of Protein IV, a 62-kDa polypeptide.29 The fiber and penton base not only contribute to the capsid structure but are intimately involved in the attachment of the viral particle to the host cell surface.
VIRAL LIFE CYCLE
The replication cycle of adenovirus may be divided into three stages, host infection, DNA replication, and construction of new viral particles. The fiber protein of the viral capsid attaches to a primary receptor on the host cell-surface, much like an anchor. Three types of host cell surface molecules can act as the primary receptor: the coxsackievirus group B and adenovirus receptor (CAR),30 the major histocompatibility complex (MHC)-I α2 subunit,31 and sialic acid residues on glycoprotein.32 Once the viral particle is tethered through the primary interaction, a second interaction between the penton base and a secondary cell-surface receptor, the integrins αvβ3 and αvβ5, take place. The integrins, in turn, promote internalization of the viral particle through receptor-mediated endocytosis.33 Once inside the cell, the outer capsid protein is partially disassembled because of acidification of the contents of the endosome. This allows the virus to escape the endosome before the formation of the lysosome34,35 and enter the cytoplasm. The process of viral internalization and release to the cytosol takes about 15 minutes.36 Virus particles are then translocated to the nucleus along the microtubule network.37 The adenoviral particle undergoes further disassembly, and, as a final step, the Ad hexon proteins remain at the nuclear membrane while the DNA is released into the nucleus.36
The stage of viral synthesis has been divided into early and late phases, the latter beginning after DNA replication.15 Host cell RNA polymerase II is the enzyme that transcribes all viral mRNA.37 Early viral mRNA synthesis has been subdivided into immediate early, delayed early, and intermediate transcription. Immediate early gene products act to regulate all subsequent early transcription.38 In addition, there is a domain of the E1A region that controls the initiation of DNA synthesis. The earliest viral protein to be expressed is E1A, which in turn induces the synthesis of other early proteins, E1B, E2, E3, and E4. Region E1B codes three polypeptides, the smallest of which, a 19-kDa protein, is the most essential.39
Region E2A codes for a single-stranded DNA binding protein of 72 kDa made in abundance in viral infected cells.40 The E2B region encodes two polypeptides, one of which is the precursor of the TP already mentioned.41 There is good evidence that the E3 region plays an important role in modulating the host response to adenoviral infection.42 The products of this region are dispensable for growth in tissue culture. However, one of its proteins, a 19-kDa glycosylated polypeptide binds to the MHC polypeptide heavy chain intracellularly and prevents it from being expressed on the cell surface. Two other proteins from the E3 region are also immunomodulatory in their function. One is instrumental in inhibiting the lysis of adenovirus-infected cells by tumor necrosis factor. The other has been shown to bind to the epidermal growth factor receptor. The E4 region produces many viral proteins, most of which have accessory roles in the viral replicative cycle.43
The intermediate transcripts code for two proteins, IX and IVa2, both of which are essential for virion assembly. Although both are synthesized early, they continue to be synthesized during and after completion of DNA synthesis. In fact, their rate of production in the late phase far exceeds that in the early phase of the replicative cycle.44
Synthesis of adenoviral DNA begins the late phase of the replicative cycle. On viral DNA synthesis, host DNA synthesis is almost completely shut down.45 Each DNA strand elongates by a continuous mechanism in the 5' to 3' direction with the pro TP molecule serving as the initial primer for DNA replication.46 Several nuclear factors have been isolated that are important in DNA replication. Nuclear factor I is a host cell protein that binds near the origin of adenoviral DNA and is essential for initiation and elongation of viral DNA synthesis.47 Nuclear factor II is a 30-kDa complex of two polypeptides48 and is necessary for full elongation of adenoviral DNA. Nuclear factor III recognizes the octamer sequence ATGCAAAT located between 36 and 54 nucleotides from the adenoviral origin of replication.49 The final factor needed for replication is termed ORP A and binds within the first 12 nucleotides of the viral genome.50 During the late phase of the replicative cycle, late mRNA synthesis begins,51,52 and host cell protein synthesis has completely shut down.53 Thus, study of viral transcription during this period has been greatly facilitated. Many of the genes transcribed during this portion of the replicative cycle code for the structural proteins already described.54 The events that control the switch from early to late viral gene expression have yet to be clearly elucidated. One hypothesis is that TP modification is instrumental in this process.55
Virion assembly is the final stage of the replicative process. Assembly begins when single polypeptides are assembled into capsomeres in the cytoplasm. Hexon monomers are assembled quickly (in only a few minutes).56 The pentons are synthesized with biphasic kinetics, one fourth are assembled rapidly in approximately 20 minutes, the rest take more than 10 hours. The isolation of temperature-sensitive mutants has led to the discovery of a 100-kDa scaffold protein extremely important in the assembly process.57 This protein is necessary for the assembly of the virion but is absent in the final product.
With the aid of temperature-sensitive mutants, several stages of virion assembly have been elucidated. The next recognizable stage of virion assembly is the light intermediate capsid stage.58 This stage consists of assembly of a structural intermediate consisting of the hexon, as well as proteins IIIa, VI, and VIII. Two proteins, 39-and 50-kDa, serve as scaffold proteins in the formation of this intermediate. A small piece of DNA from the left end of the viral genome is also associated with this intermediate.59 Next, viral DNA enters the preformed capsids through one of its vertices, and the core polypeptides are added.56
Viral DNA replication begins approximately 7 hours after infection, after which the late proteins are expressed. Viral particle assembly occurs 20 to 24 hours after infection, and approximately 104 virions per cell are released in 2 to 3 days with lysis of the host cell.