1. Neu HC. Invading organisms: Agents that kill invaders. In Wingard LR, Brody
TM, Larner J et al. (eds). Human Pharmacology: Molecular to Clinical. St
Louis: Mosby Year Book, 1991:611–764 2. Harrison JW, Svec TA. The beginning of the end of the antibiotic era? Part
I. The problem: Abuse of the “miracle drugs.” Quintessence
Int 1998;29:151 3. Demerec M. Production of Staphylococcus strains resistant to various concentrations of penicillin. Proc Acad Sci 1945;31:16 4. Demerec M. Origin of bacterial resistance to antibiotics. J Bacteriol 1948;56:63 5. Sakakibara E. Studies on the streptomycin resistant variants in bacteria. Acta
School Med Univ Kioto 1951;29:72 6. Sakakibara E. Studies on the streptomycin resistant variants in bacteria. Acta
School Med Univ Kioto 1951;29:116 7. Flaks JG, Cox EC, Witting ML et al. Polypeptide synthesis with ribosomes
from streptomycin resistant and dependent E. coli. Biochem Biophys Res Commun 1962;7:390 8. Cavalli LL, Maccacaro GA. Chloromycetin resistance in E. coli: A case of quantitative inheritance in bacteria. Nature 1950;166:991 9. Merkel JR, Steers E. Relationship between chloramphenicol reductase activity
and chloramphenicol resistance in E. coli. J Bacteriol 1953;66:389 10. Kushner DJ. The basis of chloramphenicol resistance in Pseudomonas fluorescens. Arch Biochem Biophys 1955;68:1486 11. Bannic S. Transduction to penicillin and chloramphenicol resistance in Salmonella typhimurium. Genetics 1959;44:449 12. Miyamura S. Relationship between chloramphenicol reductase activity and
chloramphenicol resistance. Jpn J Bacteriol 1961;16:115 13. Polsinelli M. Genetic analysis of oxytetracycline resistance in Bacillus subtilis by means of transformation. J Gen Microbiol 1964;36:423 14. Franklin TJ, Godfrey A. Resistance of Escherichia coli to tetracyclines. Biochem J 1965;94:54 15. Polsinelli M, Cifferi O, Cassani G et al. Mechanism of resistance to actinomycin
in Bacillus subtilis. J Bacteriol 1964;88:1567 16. Ravin AW, Iyer VN. The genetic relationship and phenotype expression of
mutations endowing Pneumococcus with resistance to erythromycin. J Gen Microbiol 1961;26:277 17. Taubman SB, Young FE, Corcoran SW. Antibiotic glycosides. IV. Studies on
the mechanism of erythromycin-resistance in Bacillus subtilis. Proc Natl Acad Sci 1963;50:955 18. Weaver JA, Patte PA. Inducible resistance to erythromycin in Staphylococcus aureus. J Bacteriol 1964;88:574 19. Foster JW, Pittillo RF. Metabolite reversal of antibiotic inhibition: Especially
reversal of aureomycin inhibition by riboflavin. J Bacteriol 1953;66:478 20. Saz AK, Martinez LM. Enzymatic basis of resistance to aureomycin. 1. J
Biol Chem 1956;223:285 21. Sutherland R, Robinson GN. Characteristics of methicillin resistant Staphylococci. J Bacteriol 1964;87:887 22. Harada K, Kameda M, Suzuki M et al. Drug resistance of enteric bacteria. II. J
Bacteriol 1964;88:1257 23. Kondo E, Mitsuhashi S. Drug resistance of enteric bacteria. III. J Bacteriol 1964;88:1261 24. Hotchkiss RD. Transfer of penicillin resistance to Pneumococcus by the DNA derived from resistant cultures. Cold Spr Harb Symp Q Biol 1957;16:45 25. Hsu YC, Herriott RM. Studies on transformations of Haemophilus influenzae. III. J Gen Physiol 1961;45:197 26. Cohen ML. Epidemiology of drug resistance: Implications for post-antimicrobial
era. Science 1992;257:1050 27. Begley S. The end of antibiotics. Newsweek 1994(Mar 28):47 28. Abbasi K. Report calls for actin on antibiotic resistance. BMJ 1998;316;1261 29. Prober CG. Bacterial resistance and the dilemma of antibiotic usage. West
J Med 1997;166:337 30. Tauxe RV, Puhr ND, Wells JG et al. Antimicrobial resistance of Shigella isolates in the USA: The importance of international travelers. J Infect
Dis 1990;162:1107 31. Appelbaum PC. Antimicrobial resistance in Streptococcus pneumoniae: An overview. Clin Infect Dis 1992;15:77 32. Bartlett JG, Frogatt JW. Antibiotic resistance. Arch Otolaryngol Head Neck
Surg 1995;121:392 33. Tomasz A. Pneumococcus at the gates. N Engl J Med 1995;333:514 34. Cunney R, Smyth E. Controlling antibiotic use: Protecting an endangered
medical resource. Ir Med J 1998;91:14 35. Arachi A. The global tuberculosis situation and the new control strategy
of the World Health Organization. Tubercle 1991;72:1 36. Bloom BR, Murray CJL. Tuberculosis: Commentary on a reemergent killer. Science 1992;257:1055 37. Barnes PF, Barrows SA. Tuberculosis in the 1990s. Ann Intern Med 1993;119:400 38. Acar JF. Consequences of bacterial resistance to antibiotics in medical
practice. Clin Infect Dis 1997;24:S17 39. Harrison JW, Svec TA. The beginning of the end of the antibiotic era? Part
II. Proposed solutions to antibiotic abuse. Quintessence Int 1998;29:223 40. Piddock LJV. Mechanisms of resistance to fluoroquinolones: State-of-the-art 1992-1994. Drugs 1995;49:29 41. Speer BS, Shoemaker NB, Salyers AA. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin Microbiol Rev 1992;5:387 42. Fralick JA. Microbial genetics. In Hentges DJ (ed). Microbiology and Immunology: An
Illustrated Review with Questions and Explanations. Boston: Little
Brown and Company, 1995:53–86 43. Holt SC, Progulske A. General microbiology, metabolism, and genetics. In
Nisengard RJ, Newman MG (eds). Oral Microbiology and Immunology. Philadelphia: Saunders, 1994:47–114 44. Borrmann LR, Leopold IH. The potential use of quinolones in future ocular
antimicrobial therapy. Am J Ophthalmol 1988;106:227 45. Gelender H, Rettich C. Gentamicin-resistant Pseudomonas aeruginosa corneal ulcers. Cornea 1984;3:21 46. Mehta NJ, Webb RM, Krohel GB et al. Clinical efficacy of tobramycin and
gentamicin sulfate in the treatment of ocular infection. Cornea 1984-1985;3:228 47. Tuft SJ, Matheson M. In vitro antibiotic resistance in bacterial keratitis
in London. Br J Ophthalmol 2000;84:687 48. Cokingtin CD, Hyndiuk RA. Insights from experimental data on ciprofloxacin
in the treatment of bacterial keratitis and ocular infections. Am
J Ophthalmol 1991;112:25S 49. Snyder ME, Katz HR. Ciprofloxacin-resistant bacterial keratitis. Am J Ophthalmol 1992;114:336 50. Garg P, Sharma S, Rao GN. Ciprofloxacin-resistant Pseudomonas keratitis. Ophthalmology 1999;106:1319 51. Goldstein MH, Kowalski RP, Gordon YJ. Emerging fluoroquinolone resistance
in bacterial keratitis: A 5-year review. Ophthalmology 1999;106:1313 52. Alexandrakis G, Alfonso EC, Miller D. Shifting trends in bacterial keratitis
in south Florida and emerging resistance to fluoroquinolones. Ophthalmology 2000;107:1497 53. Perez EM, Palma LA, Miller D et al. Penicillin resistance among ocular
pneumococcal isolates. Invest Ophthalmol Vis Sci 1996;37:S317 54. Ooishi M, Miyao M. Antibiotic sensitivity of recent clinical isolates from
patients with ocular infections. Ophthalmologica 1997;211:15–24. 55. Penland RL, Wilhelmus KR. Emergence of penicillin-resistant Streptococcus pneumoniae ocular infections. Cornea 1998;17:135 56. Jensen HG, Felix C. In vitro antibiotic susceptibilities of ocular isolates
in North and South America. Cornea 1998;17:79 57. Roth DB, Flynn HW. Antibiotic selection in the treatment of endophthalmitis: The
significance of drug combinations and synergy. Surv Ophthalmol 1997;41:395 58. Mao LK, Flynn HW, Miller D et al. Endophthalmitis caused by streptococcal
species. Arch Ophthalmol 1992;110:798 59. Endophthalmitis Vitrectomy Study Group: Results of the Endophthalmitis
Vitrectomy Study: A randomized trial of immediate vitrectomy and of intravenous
antibiotics for the treatment of postoperative bacterial endophthalmitis. Arch
Ophthalmol 1995;113:1479 60. Gordon YJ. Vancomycin prophylaxis and emerging resistance: Are ophthalmologists
the villains? The heroes? Am J Ophthalmol 2001;131:371 61. Kowalski RP, Pandya AN, Karenchak LM et al. An in vitro resistance study
of levofloxacin, ciprofloxacin, and ofloxacin using keratitis isolates
of Staphylococcus aureus and Pseudomonas aeruginosa. Ophthalmology 2001;108:1826 62. Berlin B. Antibiotics with an attitude: Are they working? N Engl J Med 1996;93:25 63. Turnidge J. Antibiotics use of misuse? Med J Aust 1997;167:116 64. Preston SL, Briceland LL. Single daily dosing of aminoglycosides. Pharmacotherapy 1995;15:297 65. Epstein JB. Infective endocarditis: Dental implications and new guidelines
for antibiotic prophylaxis. J Can Dent Assoc 1998;64:281 66. McDonald M, Grabsch E, Marshall C et al. Single- versus multiple-dose antimicrobial
prophylaxis for major surgery: A systematic review. Aust
N Z J Surg 1998;68:388 67. Glenny SF. Antimicrobial prophylaxis in colorectal surgery: A systematic
review of randomized controlled trials. Health Technol Assess 1998;2:1 68. Bogaard van den AE. Antimicrobial resistance—Relation to human and
animal exposure to antibiotics. J Antimicrob Chemother 197;40:453 69. Gerber AU, Vastola AP, Brandel J et al. Selection of aminoglycoside-resistant
variants of Pseudomonas aeruginosa in an in vivo model. J Infect Dis 1982;146:691 70. Pechere JC, Marchou B, Michea-Hamzehpour M et al. Emergence of resistance
after therapy with antibiotics used alone or combined in a murine model. J
Antimicrob Chemother 1986;17:11 71. Blaser J, Stone BB, Groner M et al. Comparative study with enoxacin and
netilmicin in a pharmacokinetic model to determine importance of ratio
of antibiotic peak concentration to MIC for bacterial activity and emergence
of resistance. Antimicrob Agents Chemother 1987;31:1054 72. Schentag JJ, Birmingham MC, Paladino JA et al. In nosocomial pneumonia, optimizing
antibiotics other than aminoglycosides is a more important
determinant of successful clinical outcome, and a better means of avoiding
resistance. Semin Respir Infect 1997;12:278 73. Jernigan DB, Cetron MS, Breiman RF. Minimizing the impact of drug-resistant Streptococcus pneumonia (DRSP). A strategy from the DRSP working group. JAMA 1996;275:206 74. McGowan JE. Minimizing antimicrobial resistance in hospital bacteria: Can
switching or cycling drugs help? J Infect Control 1986;7:573 75. Bonhoeffer S, Lipsitch M, Levin BR. Evaluating treatment protocols to prevent
antibiotic resistance. Proc Natl Acad Sci 1997;94:12106 76. Ciulla TA, Starr MB, Masket S. Bacterial endophthalmitis prophylaxis for
cataract surgery. An evidence-based update. Ophthalmology 2002;109:13 77. Swartz MN. Hospital-acquired infections: Diseases with increasingly limited
therapies. Proc Natl Acad Sci 1994;91:2420 78. Murray BE. Can antibiotic resistance be controlled? N Engl J Med 1994;330:1229 | 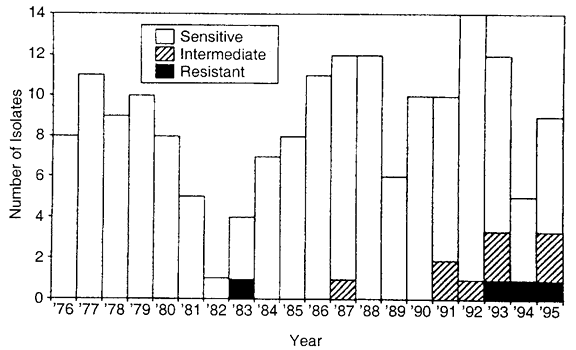
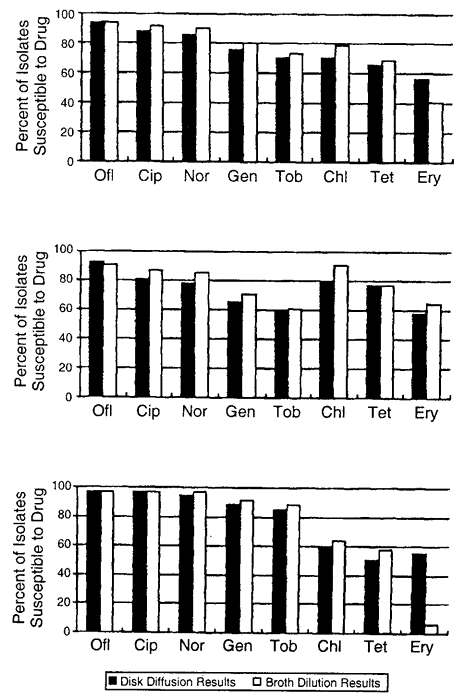
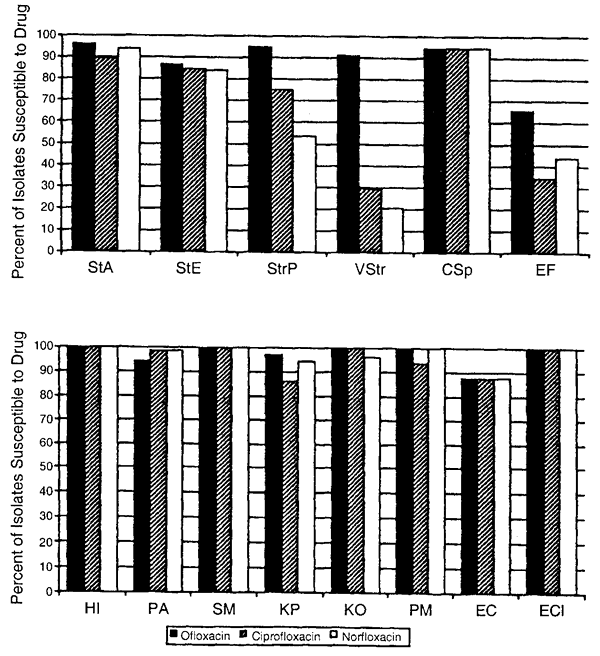
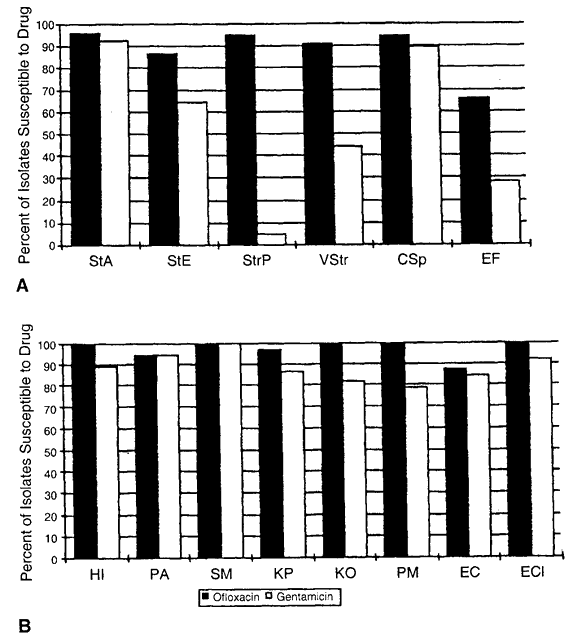
 72 hours), high dose (e.g., drops administered at least four times daily) prophylactic
regimen for many ophthalmic surgical procedures,
72 hours), high dose (e.g., drops administered at least four times daily) prophylactic
regimen for many ophthalmic surgical procedures,