
|
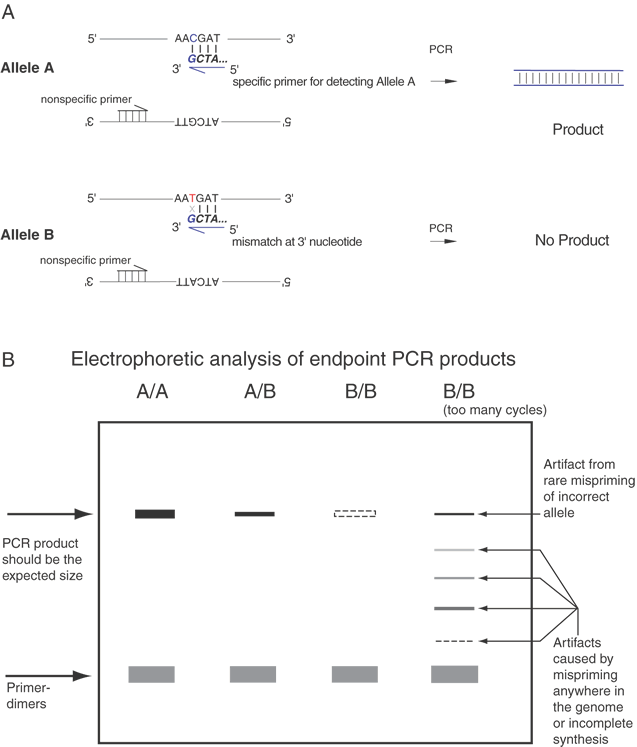
| Fig. 11. Allele-specific PCR by endpoint analysis. Panel A. Allele A and Allele B can be distinguished by PCR assay provided that an allele-specific primer (with a perfect match to one allele and a single base mismatch at the other allele) is known. In the illustrated case, a 3' terminal G matches perfectly to a C in the upper strand of Allele A, and the 3' G mismatches with a T on the upper strand of Allele B. Under optimized conditions for the PCR, the reaction can be tuned to produce large amounts of PCR product from allele A with little or none from Allele B. Panel B. Electrophoretic analysis of PCR products from individuals who are homozygous for Allele A (A/A), heterozygous for Alleles A and B (A/B), and homozygous for Allele B (B/B). A heavy PCR band is expected for any A/A individuals, about half as much product is expected for heterozygous (A/B) individuals, and no product is expected from homozygous (B/B) individuals. The electrophoretic gel analysis allows the measurement of the size of the PCR product, and it should be the expected size. The product can be restriction digested to test whether the product has the anticipated restriction enzyme sites, and the product can be sequenced to further validate the its. Several possible artifacts of the PCR process are illustrated in Panel B. Primer-dimers can occur if the primers can bind to each other. Because of the high concentrations of the primers in the PCR, there is a propensity for the primers to bind to each other even when just few bases near the 3' ends of the primers match each other. The primer dimers are detected near the bottom of the gel corresponding to the sum of the lengths of the primers, usually about 40 base pairs. Increasing the annealing temperature and decreasing the concentration of the primers may reduce the amount of primer-dimers. The far right lane [labeled B/B (too many cycles)] illustrates several other potential PCR artifacts. These are usually most noticeable when the PCR is subjected to too many cycles. In most cases, with 10 ng genomic DNA, a PCR is complete by 25 to 30 cycles. In certain circumstances it may be necessary to run the reaction for more cycles, and under those conditions a series of artifacts may be detected. One artifact is that after too many cycles, reagents and substrates will become rate limiting and products may not be completely copied. These partial products can be detected at many intermediate sizes, from very small to nearly full size. Also, after being subjected to repeated heating, contaminants in the reagents may reduce the specificity of the annealing, and mispriming can occur. This may lead to the formation of a full-ength PCR product, even in the presence of only Allele B DNA. Thus, the PCR must be optimized for the number of cycles to maximize the yield of the A/A product, minimize the amount of any B/B product, minimize primer-dimers, and minimize any incomplete products. The optimization process usually titrates the number of PCR cycles, annealing temperature, concentrations of primers, Mg++ ions, buffers, and type and amount of polymerase. Usually, several candidate primer pairs are tested. |